Filter by OSSI funding status
 Scientific Computing Software Turaga Lab Python GPL-3.0 Package Single-molecule localization microscopy (SMLM) Google Colab Jupyter notebook Local installation PyTorch
Scientific Computing Software Turaga Lab Python GPL-3.0 Package Single-molecule localization microscopy (SMLM) Google Colab Jupyter notebook Local installation PyTorch An open-source platform for single-molecule localization
Develop a user-friendly (napari) deep learning-based single-molecule detection software.
 Scientific Computing Software Java BSD-2 Clause GPL-2.0 Command line application Expansion microscopy (ExM) Image registration Lightsheet fluorescence microscopy (LFSM) Spatial transcriptomics Cloud HPC cluster Local installation BigDataViewer ImgLib2 Bioformats compatible formats HDF5 N5 TIFF ZARR
Scientific Computing Software Java BSD-2 Clause GPL-2.0 Command line application Expansion microscopy (ExM) Image registration Lightsheet fluorescence microscopy (LFSM) Spatial transcriptomics Cloud HPC cluster Local installation BigDataViewer ImgLib2 Bioformats compatible formats HDF5 N5 TIFF ZARR BigStitcher-Spark
Run functionality of BigStitcher distributed on your workstation, a cluster or the cloud using Apache Spark.
 Scientific Computing Software Java BSD-3 Clause Package Expansion microscopy (ExM) Image registration Lightsheet fluorescence microscopy (LFSM) Spatial transcriptomics Local installation BigDataViewer Fiji ImgLib2 Bioformats compatible formats HDF5 N5 TIFF ZARR Zeiss CZI
Scientific Computing Software Java BSD-3 Clause Package Expansion microscopy (ExM) Image registration Lightsheet fluorescence microscopy (LFSM) Spatial transcriptomics Local installation BigDataViewer Fiji ImgLib2 Bioformats compatible formats HDF5 N5 TIFF ZARR Zeiss CZI BigStitcher
A software package that allows simple and efficient alignment of multi-tile and multi-angle image datasets, for example acquired by lightsheet, widefield or confocal microscopes.
 Ahrens Lab MultiFISH Scientific Computing Software C++ Python BSD-3 Clause Framework Package Annotation Calcium imaging Confocal light microscopy (LM) Correlative light EM (CLEM) Electron microscopy (EM) Expansion microscopy (ExM) Image registration Lightsheet fluorescence microscopy (LFSM) Single-molecule localization microscopy (SMLM) Spatial transcriptomics Cloud HPC cluster Jupyter notebook Local installation N5 OME-Zarr TIFF Zeiss CZI
Ahrens Lab MultiFISH Scientific Computing Software C++ Python BSD-3 Clause Framework Package Annotation Calcium imaging Confocal light microscopy (LM) Correlative light EM (CLEM) Electron microscopy (EM) Expansion microscopy (ExM) Image registration Lightsheet fluorescence microscopy (LFSM) Single-molecule localization microscopy (SMLM) Spatial transcriptomics Cloud HPC cluster Jupyter notebook Local installation N5 OME-Zarr TIFF Zeiss CZI BigStream
Flexible and powerful image registration tools for small and big data (TB+) alike. Stitching, subject-to-atlas, multi-modal, in vivo to ex vivo, and motion correction. 100% Python, can be run from Jupyter notebooks, including full access to workstation or adaptable distributed resources.
Catena
Pipeline for automated connectome reconstruction from volume Electron Microscopy.
 Saalfeld Lab Java GPL-2.0 Native application Confocal light microscopy (LM) Correlative light EM (CLEM) Electron microscopy (EM) Expansion microscopy (ExM) Image registration Local installation BigDataViewer Fiji ImgLib2 Java Virtual Machine HDF5 N5 OME-Zarr TIFF Zeiss CZI
Saalfeld Lab Java GPL-2.0 Native application Confocal light microscopy (LM) Correlative light EM (CLEM) Electron microscopy (EM) Expansion microscopy (ExM) Image registration Local installation BigDataViewer Fiji ImgLib2 Java Virtual Machine HDF5 N5 OME-Zarr TIFF Zeiss CZI BigWarp
Maintain and extend BigWarp (3D non-rigid registration for very large volumes).
 Pachitariu Lab Stringer Lab Python BSD-3 Clause GPL-2.0 Package Calcium imaging Confocal light microscopy (LM) Electron microscopy (EM) Expansion microscopy (ExM) Google Colab Local installation Napari
Pachitariu Lab Stringer Lab Python BSD-3 Clause GPL-2.0 Package Calcium imaging Confocal light microscopy (LM) Electron microscopy (EM) Expansion microscopy (ExM) Google Colab Local installation Napari Cellpose
A generalist algorithm for cell and nucleus segmentation (v1.0) that can be optimized for your own data (v2.0) and perform image restoration (v3.0).
DaCapo
A framework for easy application of established machine learning techniques on large, multi-dimensional images.
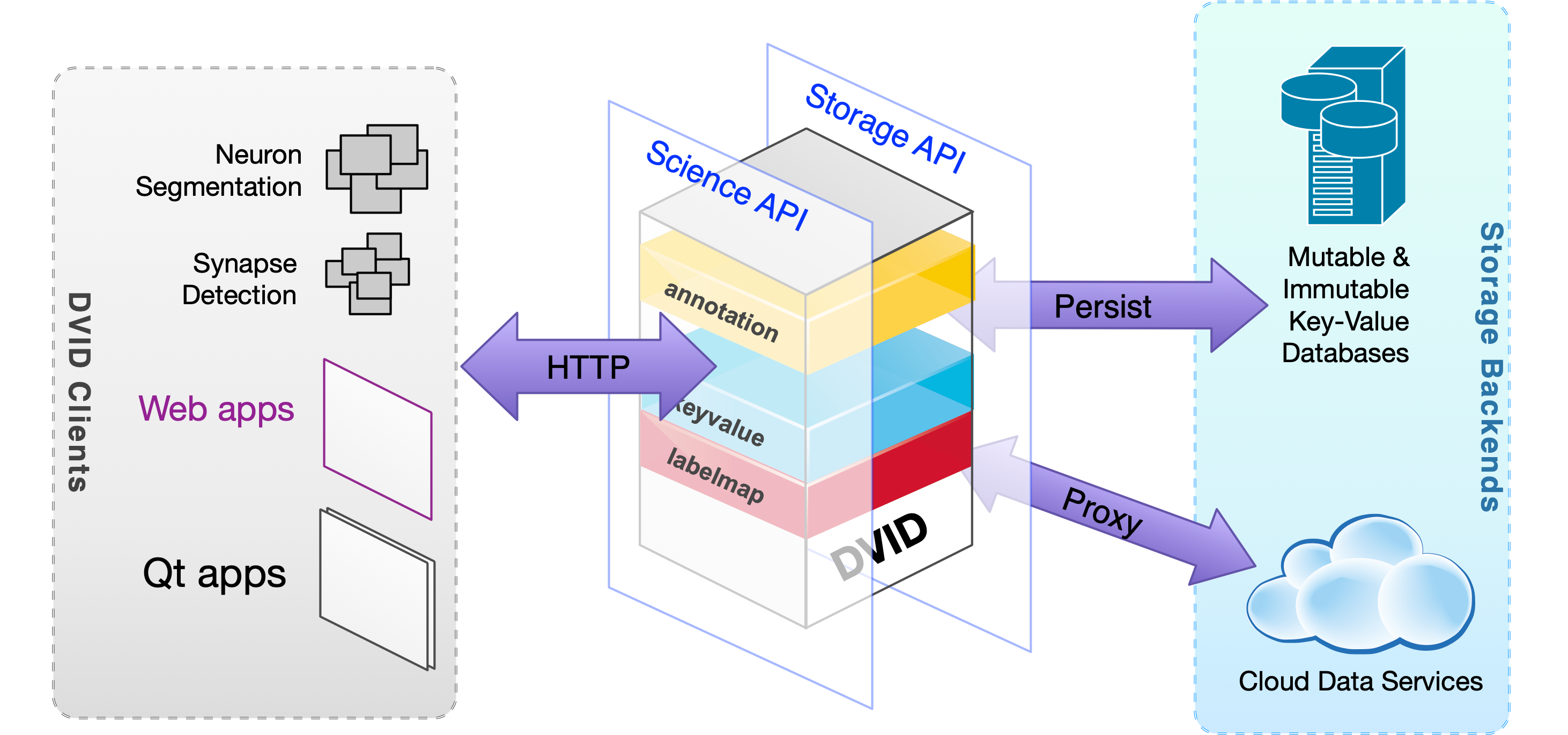 FlyEM Go BSD-3 Clause Command line application Framework Native application Service Web application Annotation Confocal light microscopy (LM) Connectomics Data service Electron microscopy (EM) Expansion microscopy (ExM) Segmentation Cloud Local installation Neuroglancer Neuroglancer precomputed
FlyEM Go BSD-3 Clause Command line application Framework Native application Service Web application Annotation Confocal light microscopy (LM) Connectomics Data service Electron microscopy (EM) Expansion microscopy (ExM) Segmentation Cloud Local installation Neuroglancer Neuroglancer precomputed DVID
A dataservice for branched versioning of a variety of data types including teravoxel-scale image volumes, sparse volumes, meshes, and JSON and point annotations.
 MultiFISH Scientific Computing Software Java MATLAB Nextflow Python BSD-3 Clause Command line application Expansion microscopy (ExM) Spatial transcriptomics Cloud HPC cluster Local installation BigDataViewer N5 Zeiss CZI
MultiFISH Scientific Computing Software Java MATLAB Nextflow Python BSD-3 Clause Command line application Expansion microscopy (ExM) Spatial transcriptomics Cloud HPC cluster Local installation BigDataViewer N5 Zeiss CZI EASI-FISH Pipeline
Automated analysis pipeline for EASI-FISH spatial transcriptomics data
Entrypoints
Support multiple entrypoint scripts in a container for use in containerized scientific tools.
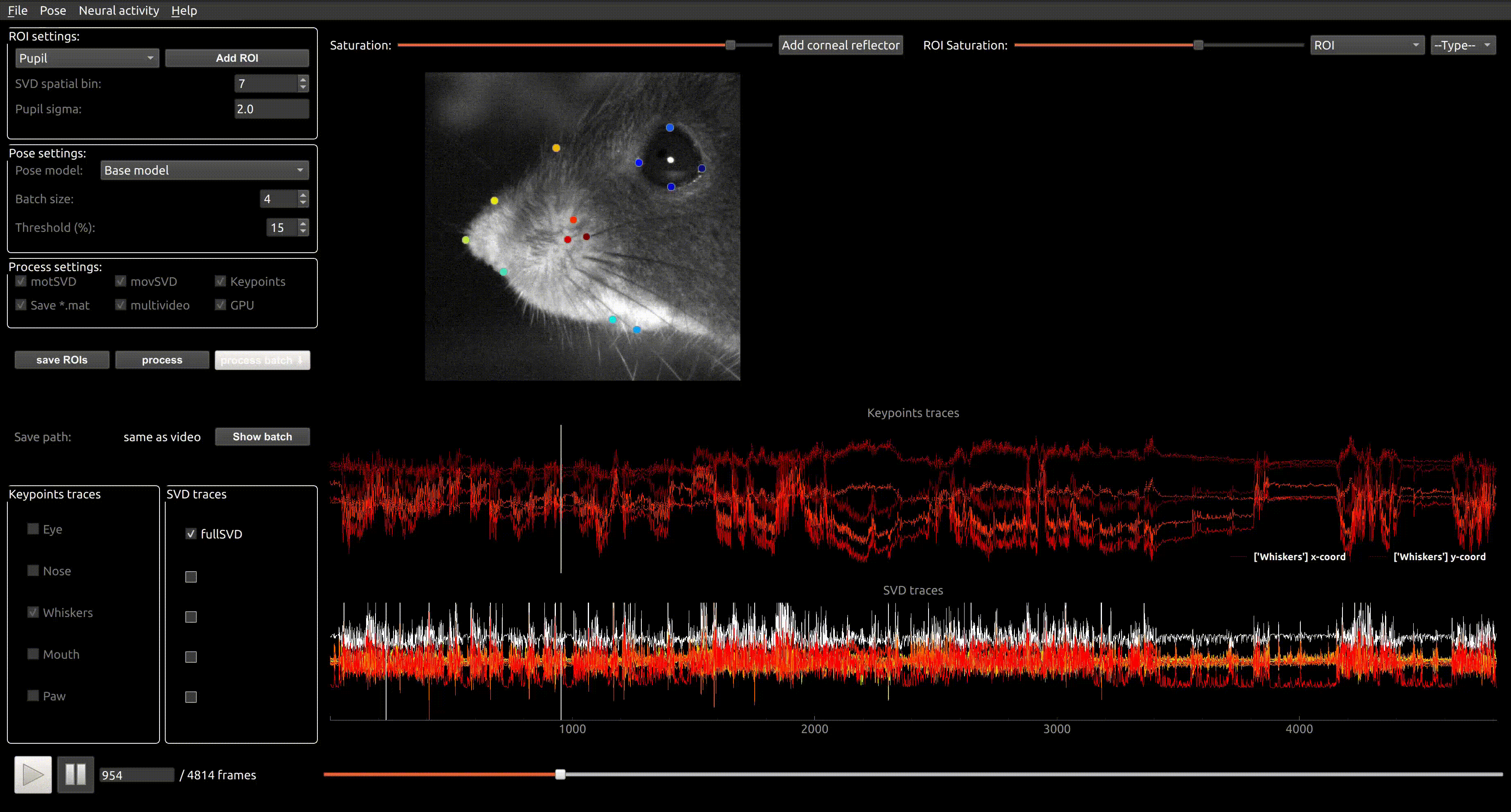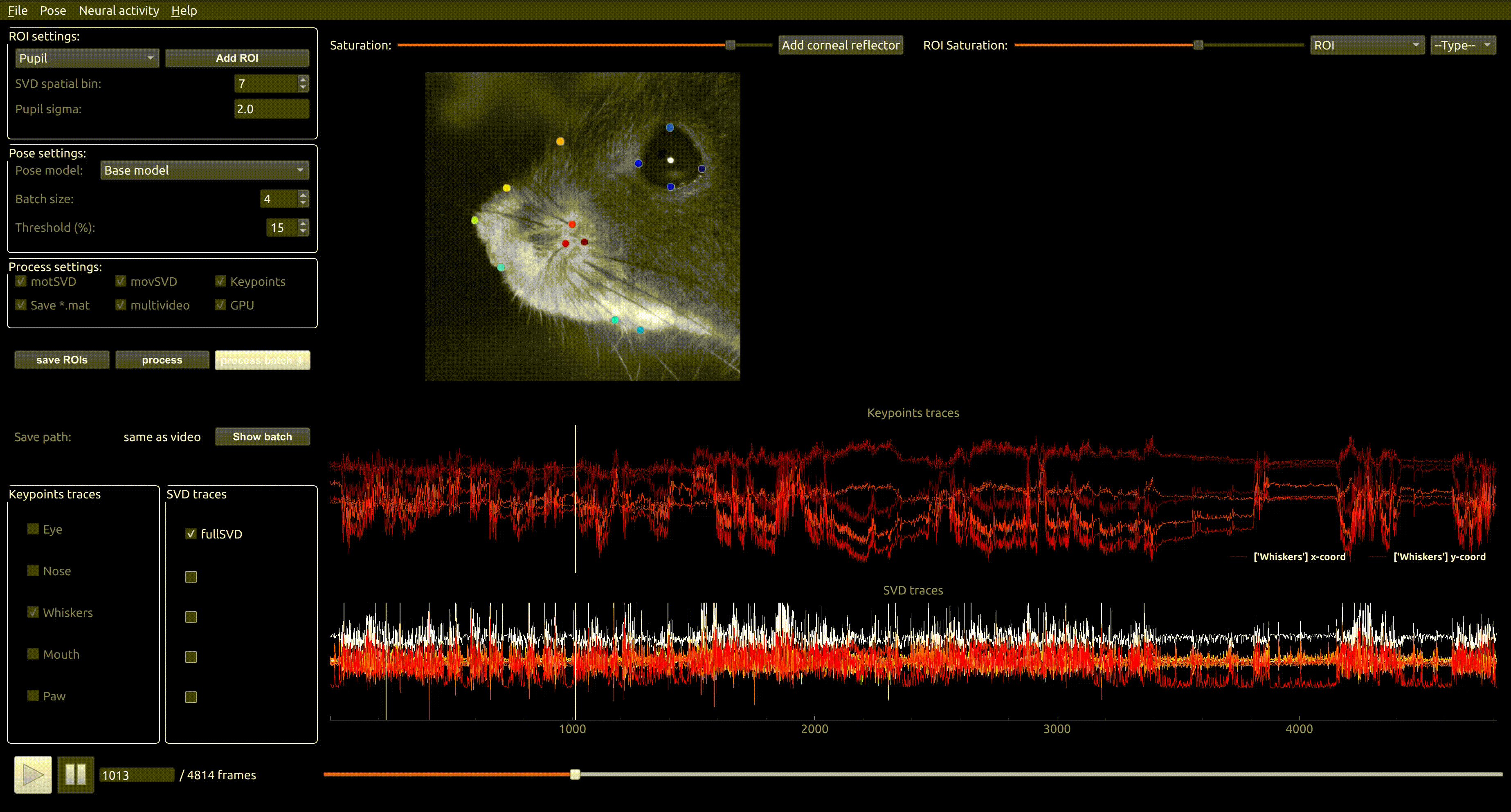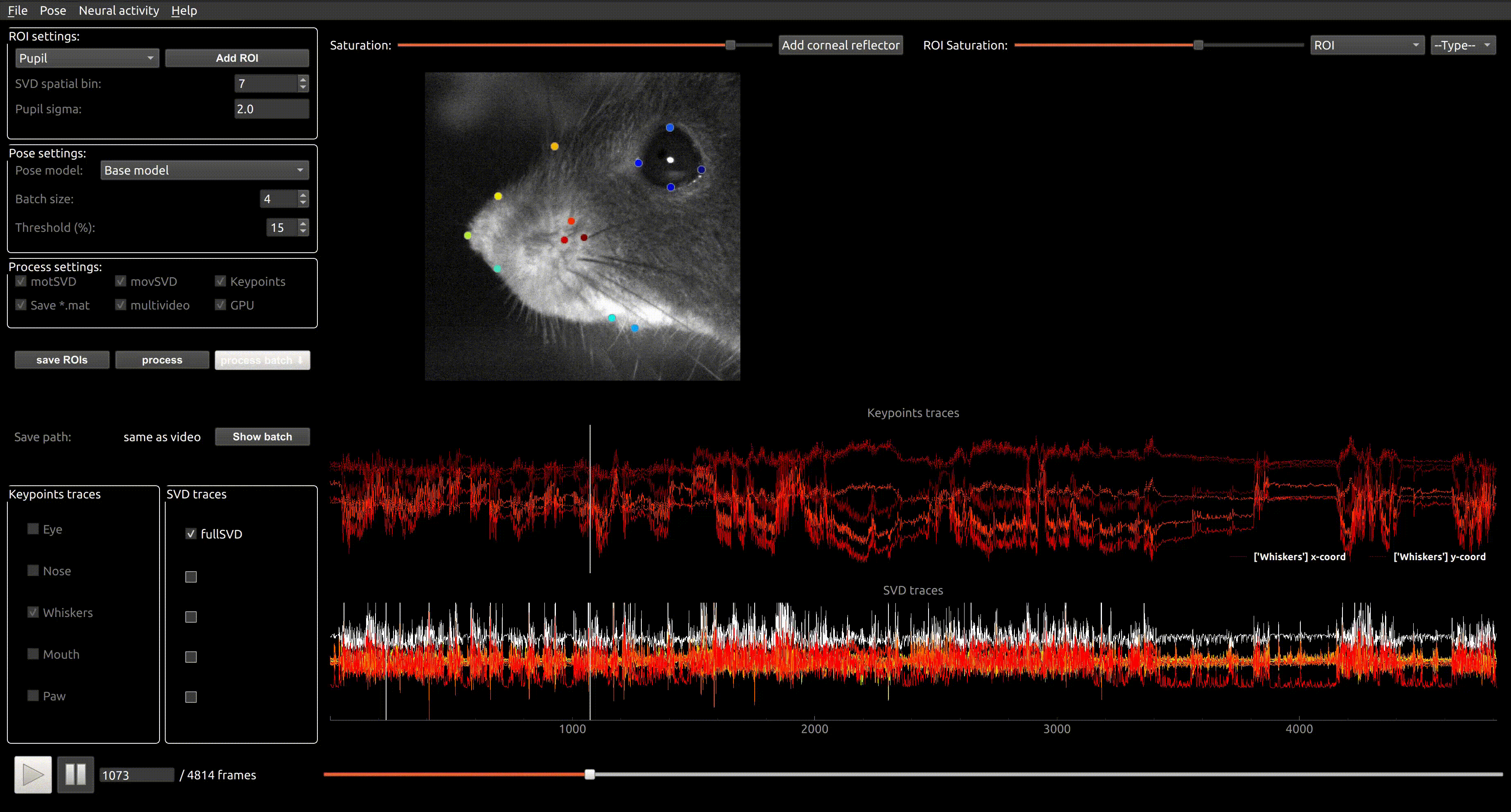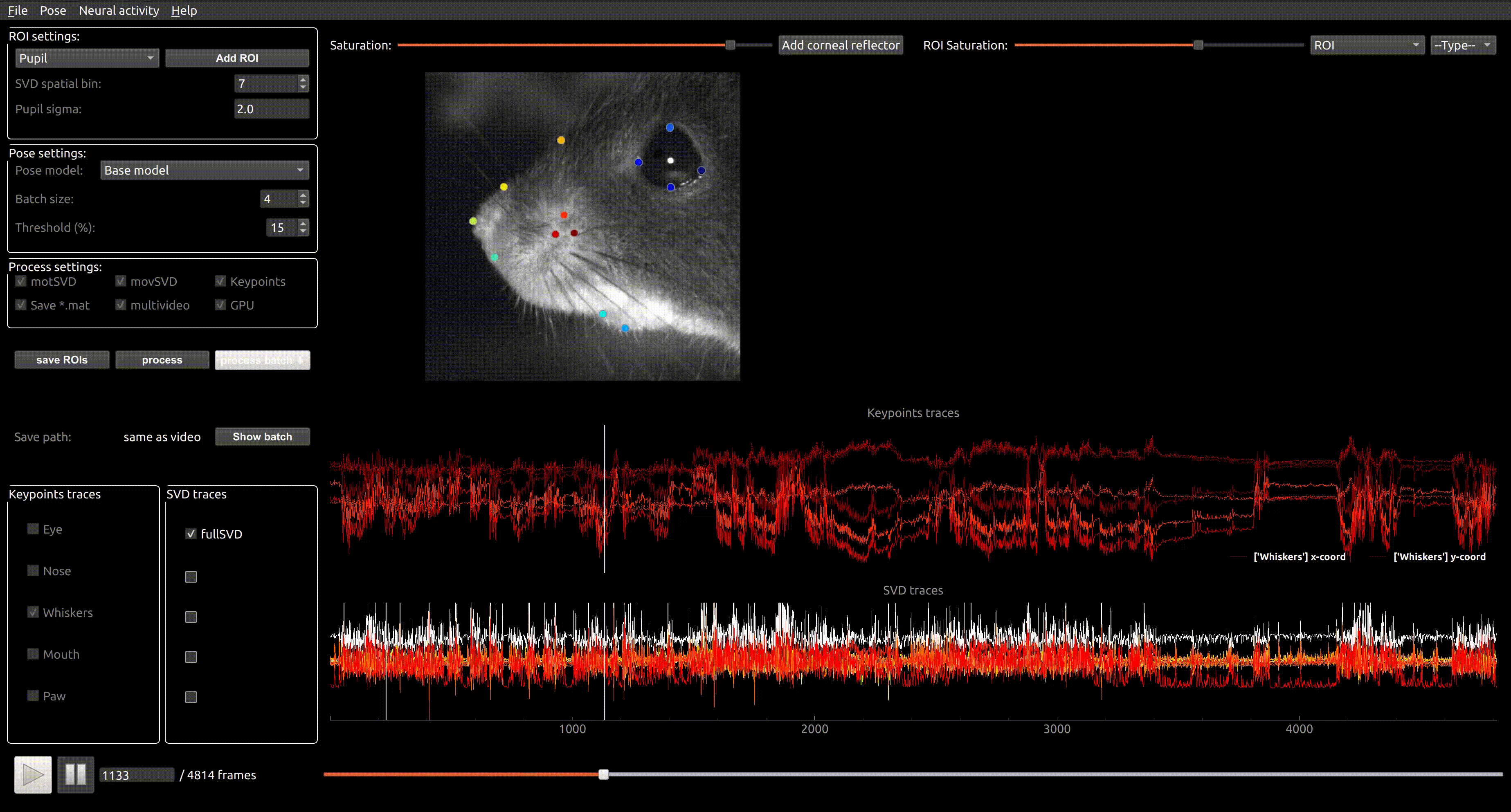 Pachitariu Lab Stringer Lab Python GPL-3.0 Command line application GUI application Package Neural recording Google Colab Jupyter notebook Local installation avi mpeg
Pachitariu Lab Stringer Lab Python GPL-3.0 Command line application GUI application Package Neural recording Google Colab Jupyter notebook Local installation avi mpeg Facemap
A framework for predicting neural activity from mouse orofacial movements. It includes a pose estimation model for tracking distinct keypoints on the mouse face, a neural network model for predicting neural activity using the pose estimates, and also can be used compute the singular value decomposition (SVD) of behavioral videos.
 Funke Lab Live Image Tracking Tools Java Python BSD-3 Clause Package Tracking Local installation Fiji Napari N5 OME-Zarr ZARR
Funke Lab Live Image Tracking Tools Java Python BSD-3 Clause Package Tracking Local installation Fiji Napari N5 OME-Zarr ZARR GEFF - Graph Exchange File Format
A Python and Java compatible file format for exchanging graphs with annotated nodes and edges, with special support for tracking graphs.
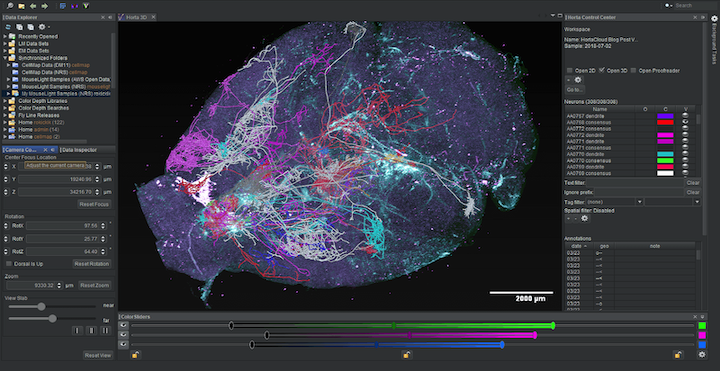 MouseLight Scientific Computing Software Java Typescript BSD-3 Clause Native application Service Web application Website Annotation Expansion microscopy (ExM) Lightsheet fluorescence microscopy (LFSM) Cloud Web browser Janelia Workstation Java Virtual Machine OME-Zarr SWC
MouseLight Scientific Computing Software Java Typescript BSD-3 Clause Native application Service Web application Website Annotation Expansion microscopy (ExM) Lightsheet fluorescence microscopy (LFSM) Cloud Web browser Janelia Workstation Java Virtual Machine OME-Zarr SWC HortaCloud
Large volume volumetric rendering and collaborative neuron annotation in the cloud
Java packaging tool chain
Develop an easy-to-use packaging system for Java applications.
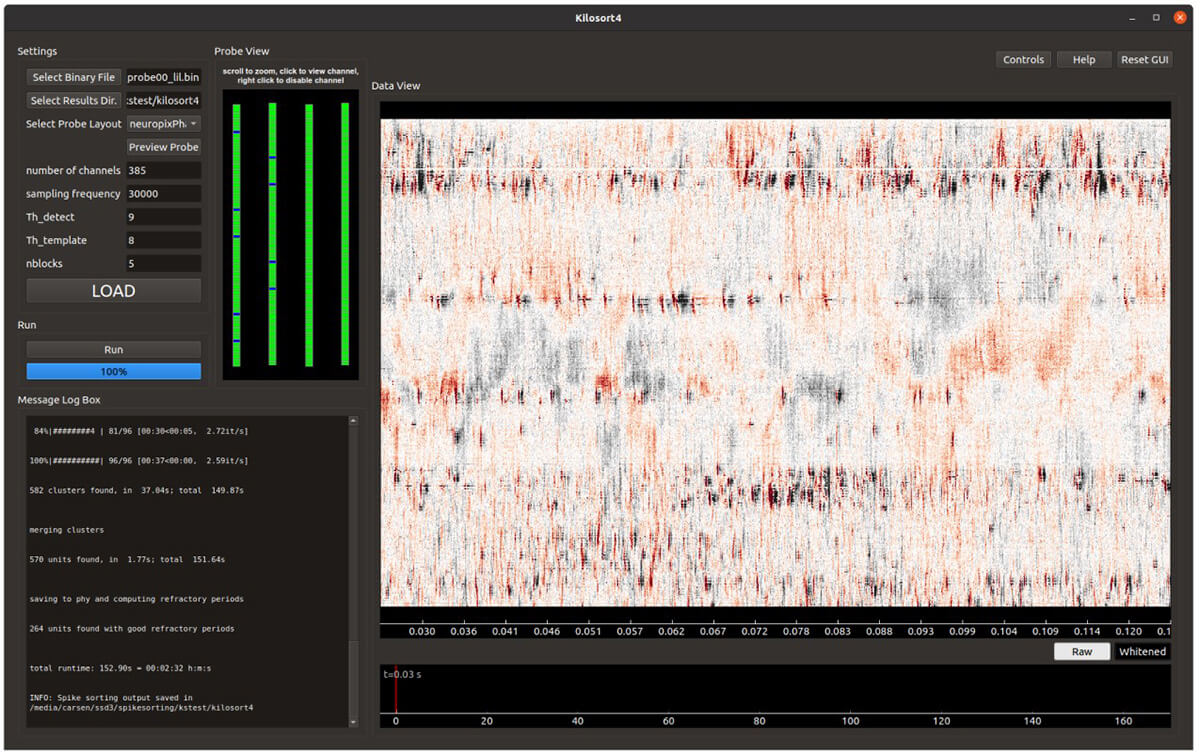 Pachitariu Lab Stringer Lab MATLAB Python GPL-2.0 Package Electrophysiology Neural recording Google Colab Local installation
Pachitariu Lab Stringer Lab MATLAB Python GPL-2.0 Package Electrophysiology Neural recording Google Colab Local installation Kilosort
Maintain and develop python version of Kilosort (electrophysiology data).
Maru
Maru is an opinionated command-line interface for quickly and easily containerizing scientific applications.
Motile Tracker
An application for interactive tracking with motile. Motile is a library that makes it easy to solve tracking problems using optimization by framing the task as an Integer Linear Program (ILP).
 Scientific Computing Software Java Javascript Python BSD-3 Clause Service Web application Website Confocal light microscopy (LM) Electron microscopy (EM) Cloud Web browser H5J SWC
Scientific Computing Software Java Javascript Python BSD-3 Clause Service Web application Website Confocal light microscopy (LM) Electron microscopy (EM) Cloud Web browser H5J SWC NeuronBridge
A web application for easily and rapidly finding putative morphological matches between large data sets of neurons imaged using different modalities, namely electron microscopy (EM) and light microscopy (LM). Matches have been precomputed for all of Janelia's public EM/LM data sets, and are quick to look up by identifier. You can also upload your own data and match it against these public data sets.
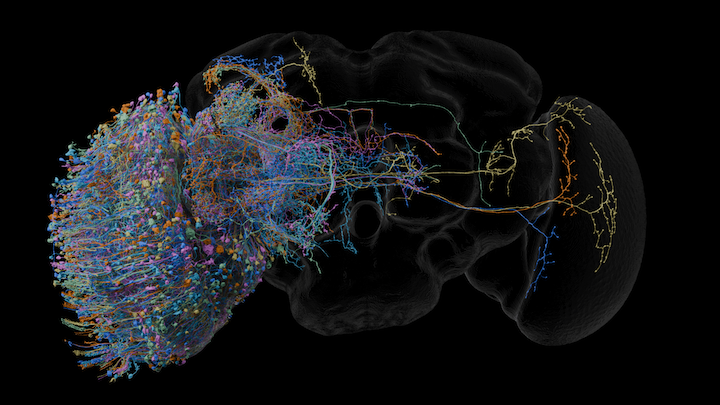 FlyEM Scientific Computing Software Python CC-by-0 Command line application Native application Confocal light microscopy (LM) Electron microscopy (EM) Visualization Local installation Blender H5J OBJ SWC
FlyEM Scientific Computing Software Python CC-by-0 Command line application Native application Confocal light microscopy (LM) Electron microscopy (EM) Visualization Local installation Blender H5J OBJ SWC neuVid
Generates biology videos from high-level descriptions using Blender or VVD Viewer.
 Scientific Computing Software Julia BSD-3 Clause Package Local installation
Scientific Computing Software Julia BSD-3 Clause Package Local installation NIDAQ.jl
A Julia wrapper for National Instruments' driver
OSSI Website
A portfolio for projects supported by Open Science Software Initiative, and other software projects at Janelia
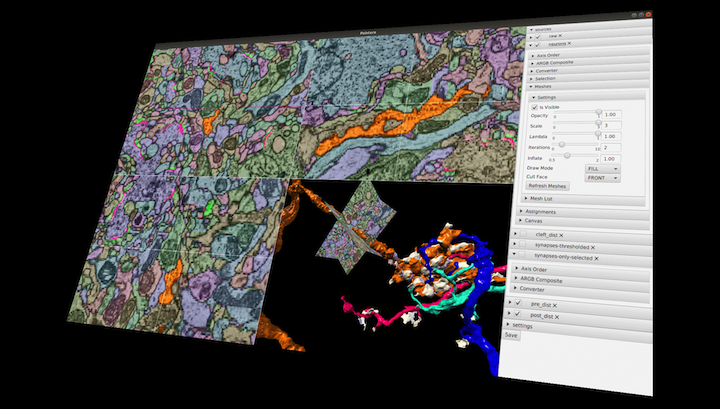 Saalfeld Lab Java Kotlin GPL-2.0 Native application Annotation Electron microscopy (EM) Visualization Local installation BigDataViewer ImgLib2 N5
Saalfeld Lab Java Kotlin GPL-2.0 Native application Annotation Electron microscopy (EM) Visualization Local installation BigDataViewer ImgLib2 N5 Paintera
Annotation and Visualization of Large 3D Datasets with Paintera
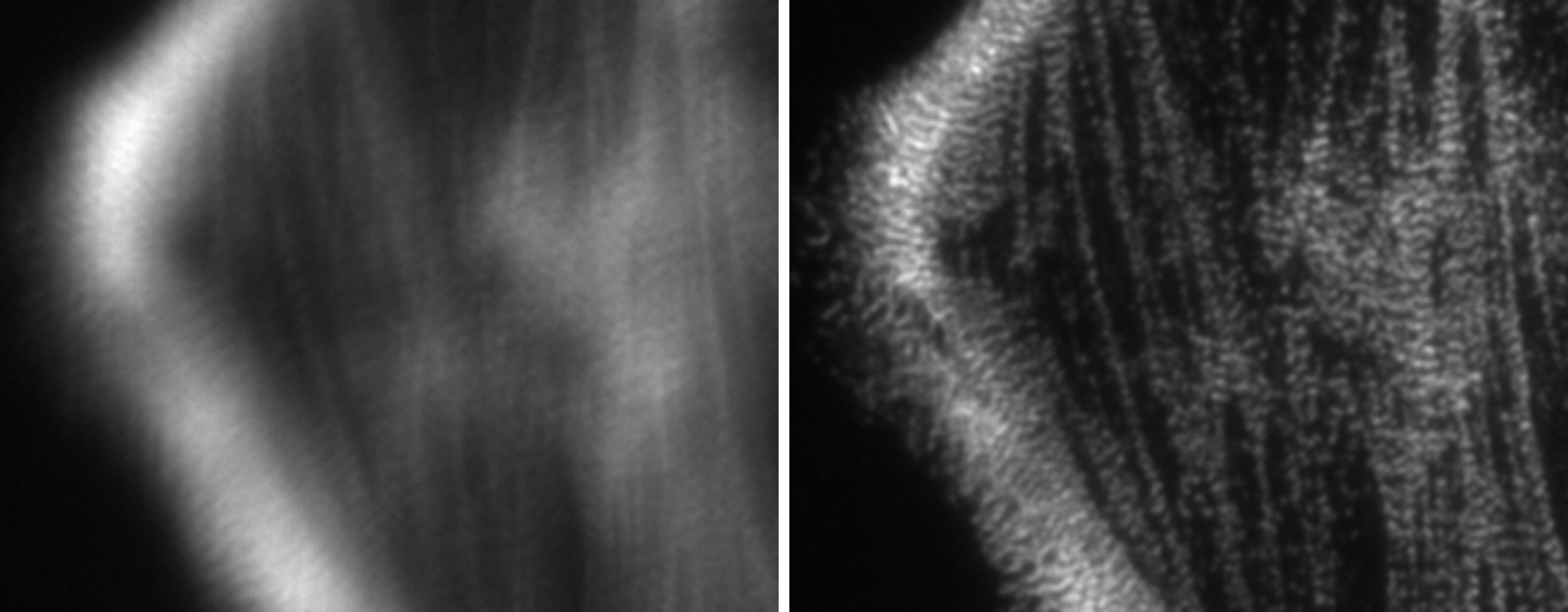 Shroff Lab MATLAB CC-by-4 Package Fluorescence microscopy Local installation TIFF
Shroff Lab MATLAB CC-by-4 Package Fluorescence microscopy Local installation TIFF Phase Diversity
Software for analyzing phase diversity data for adaptive optics
 Pachitariu Lab Stringer Lab Python GPL-3.0 Command line application GUI application Package Annotation Electrophysiology Neural recording Visualization Jupyter notebook Local installation
Pachitariu Lab Stringer Lab Python GPL-3.0 Command line application GUI application Package Annotation Electrophysiology Neural recording Visualization Jupyter notebook Local installation rastermap
A visualization method for neural data
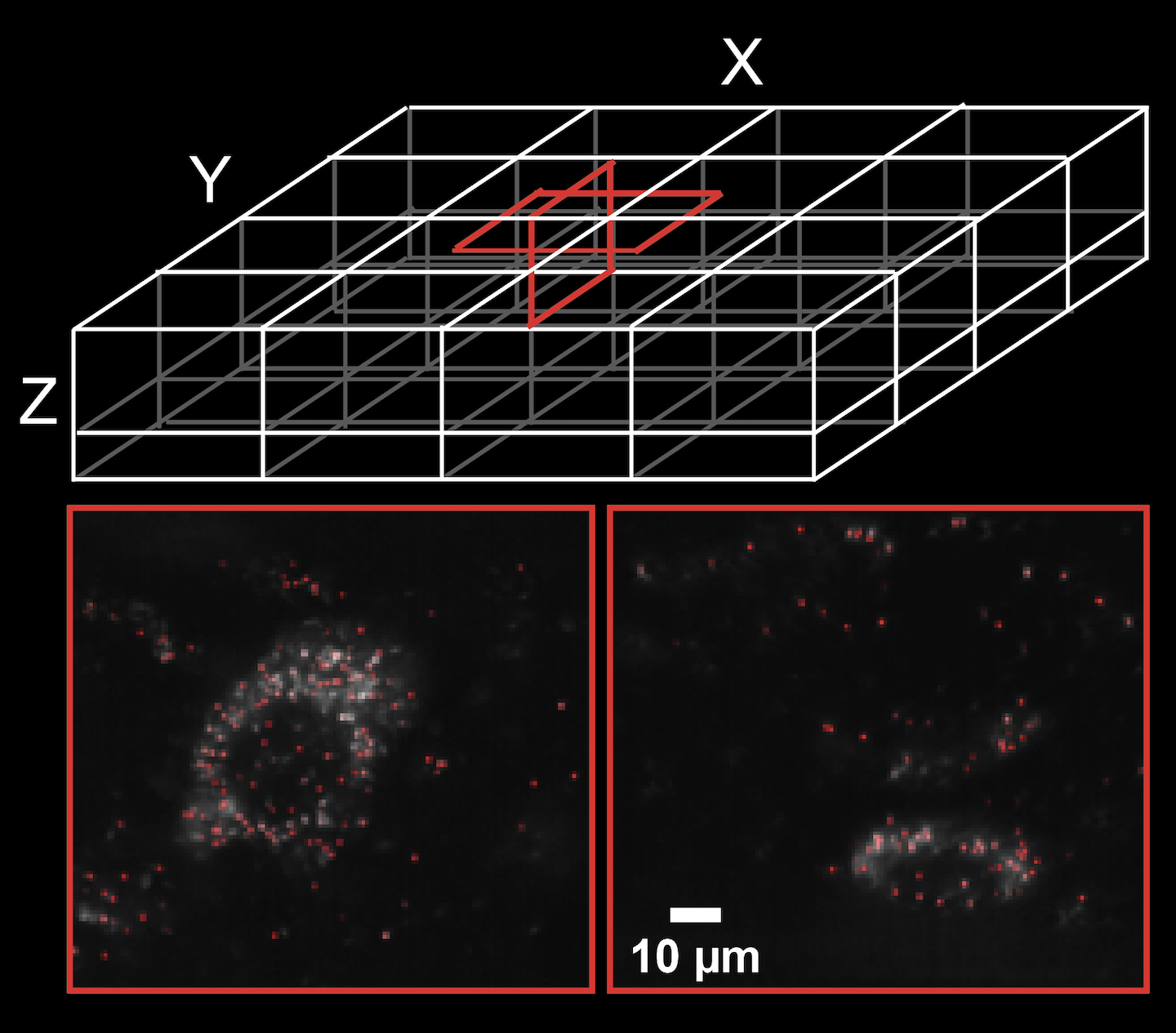 Scientific Computing Software Java GPL-2.0 GPL-3.0 Command line application Fiji plugin Framework GUI application Sequence analysis Spatial transcriptomics Cloud HPC cluster Local installation BigDataViewer Fiji ImgLib2 Spark Bioformats compatible formats N5 ZARR
Scientific Computing Software Java GPL-2.0 GPL-3.0 Command line application Fiji plugin Framework GUI application Sequence analysis Spatial transcriptomics Cloud HPC cluster Local installation BigDataViewer Fiji ImgLib2 Spark Bioformats compatible formats N5 ZARR RS-FISH
Precise, interactive, fast, and scalable FISH spot detection
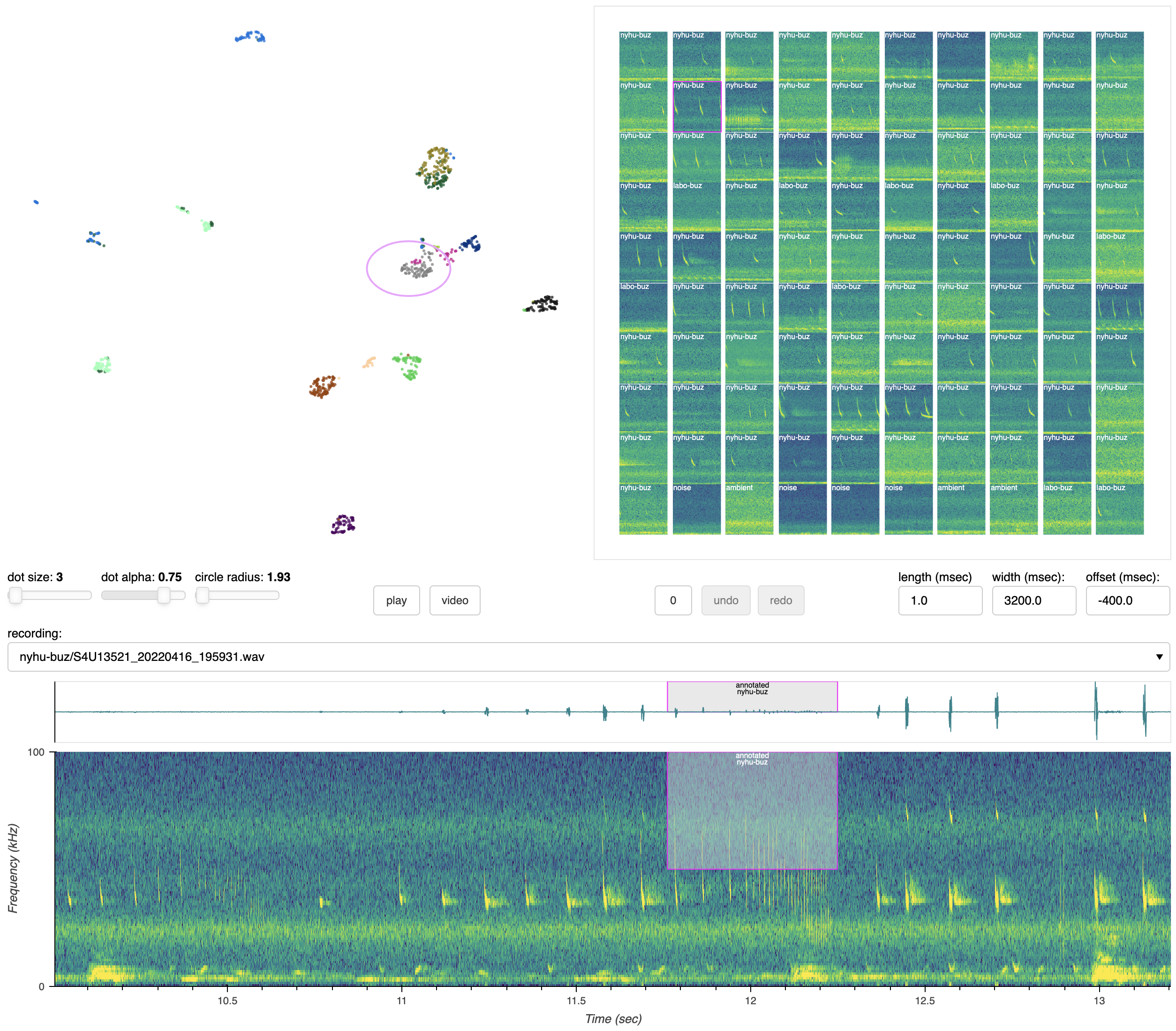 Scientific Computing Software Python BSD-3 Clause GUI application Annotation Segmentation HPC cluster Local installation Tensorflow wav
Scientific Computing Software Python BSD-3 Clause GUI application Annotation Segmentation HPC cluster Local installation Tensorflow wav SongExplorer
deep learning for acoustic signals
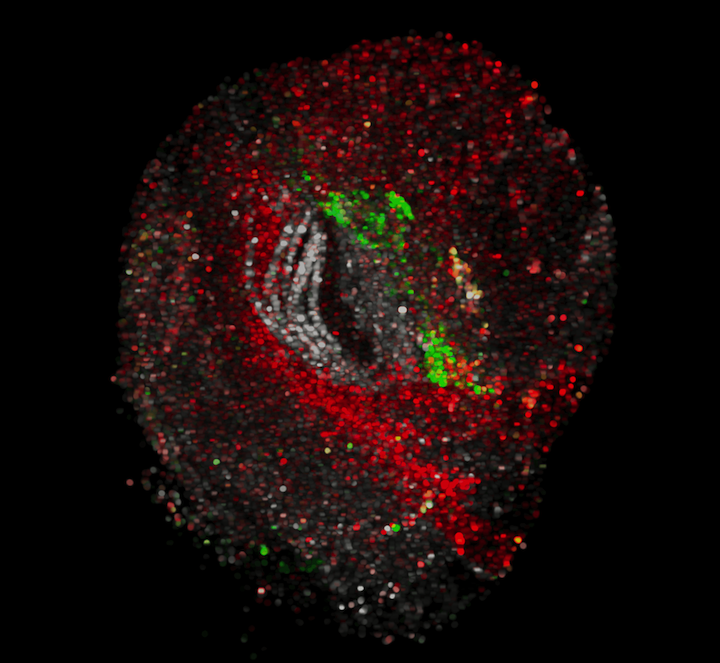 Scientific Computing Software Java GPL-3.0 Command line application Framework Native application Sequence analysis Spatial transcriptomics Local installation BigDataViewer Fiji ImgLib2 AnnData N5 Text files
Scientific Computing Software Java GPL-3.0 Command line application Framework Native application Sequence analysis Spatial transcriptomics Local installation BigDataViewer Fiji ImgLib2 AnnData N5 Text files Spatial Transcriptomics as Images Project (STIM)
A framework for storing, (interactively) viewing, and aligning spatial transcriptomics data.
Stitching-Spark
Reconstructing large microscopy images from overlapping image tiles on a high-performance Spark cluster.
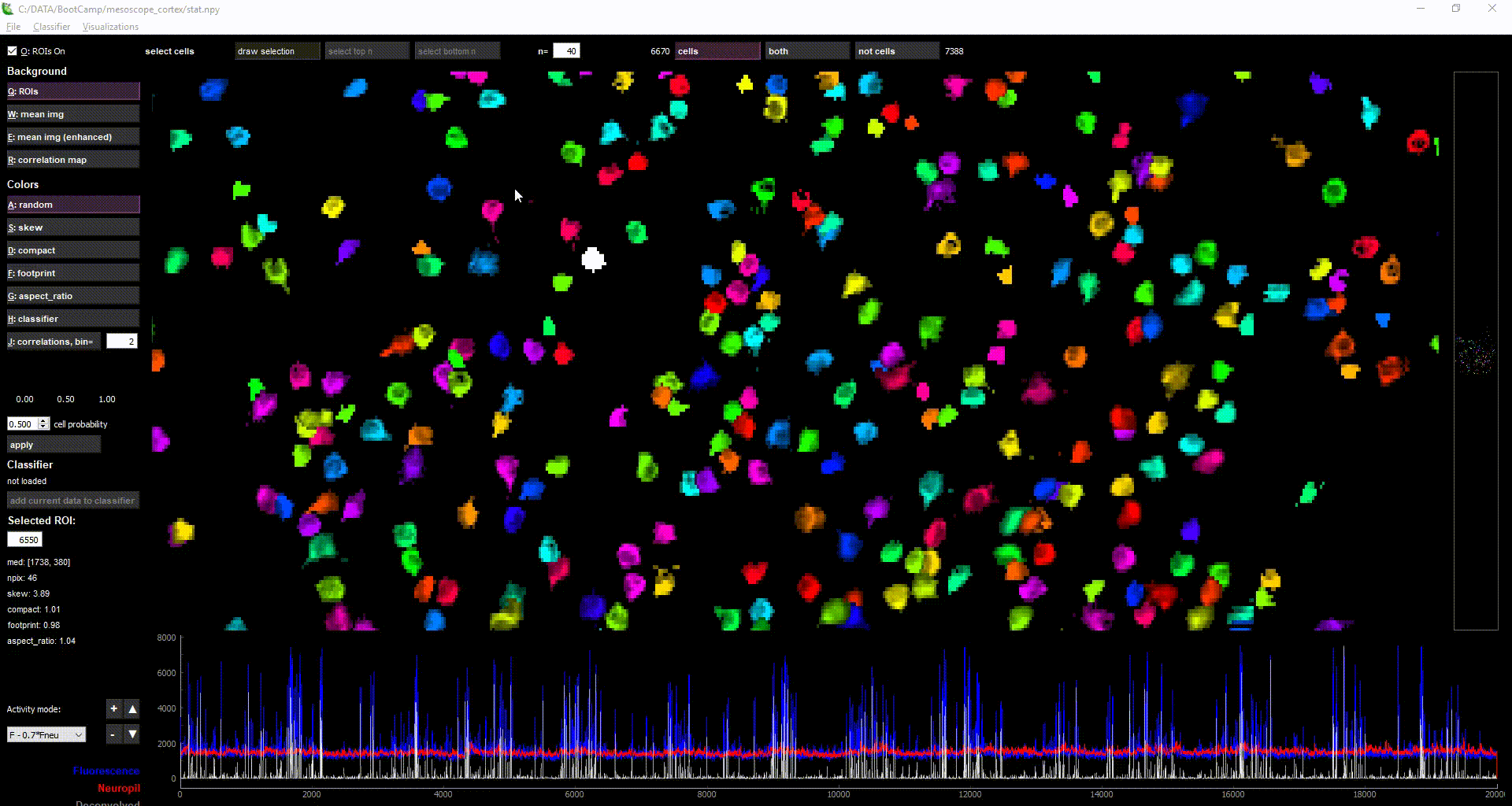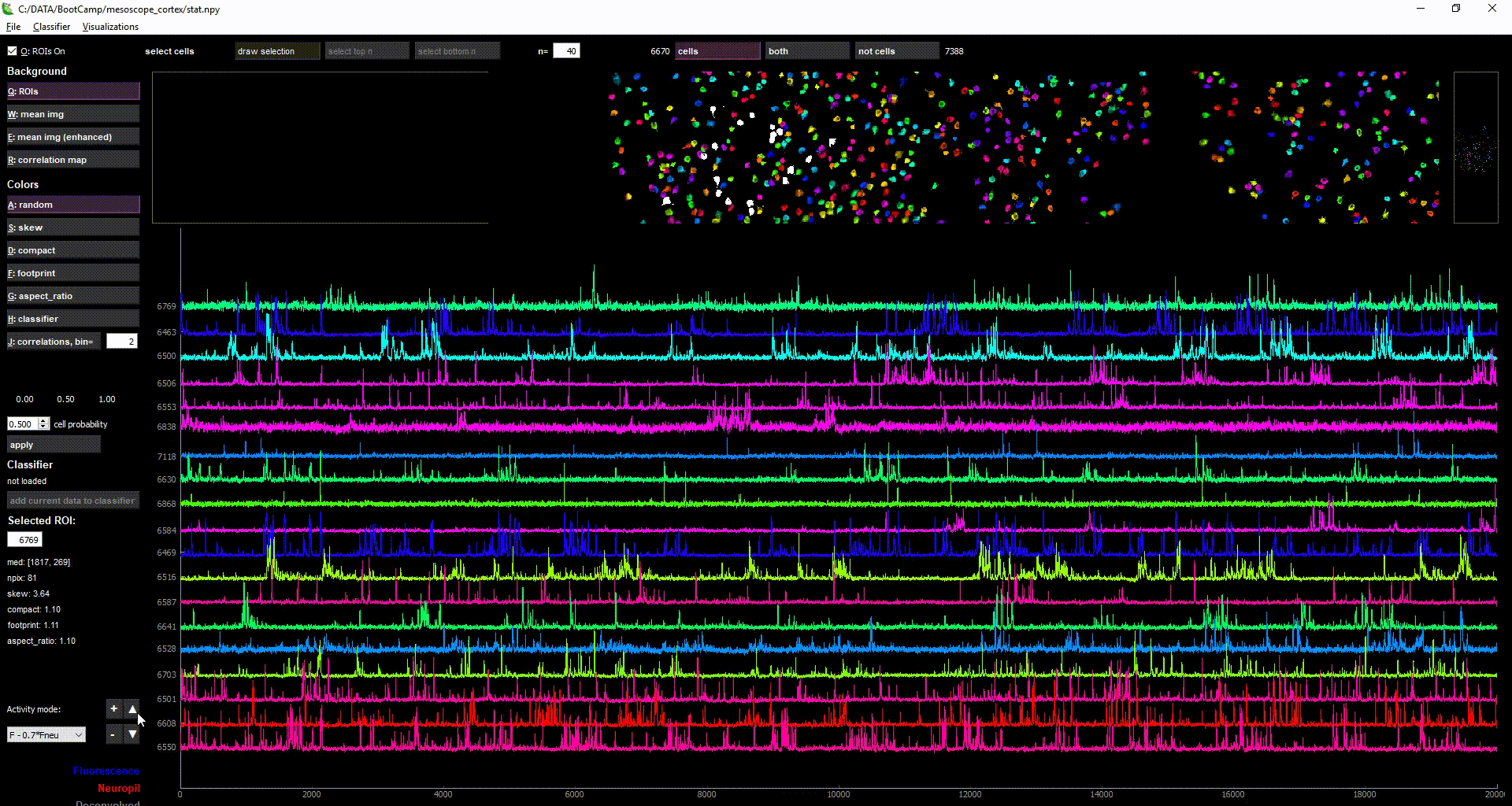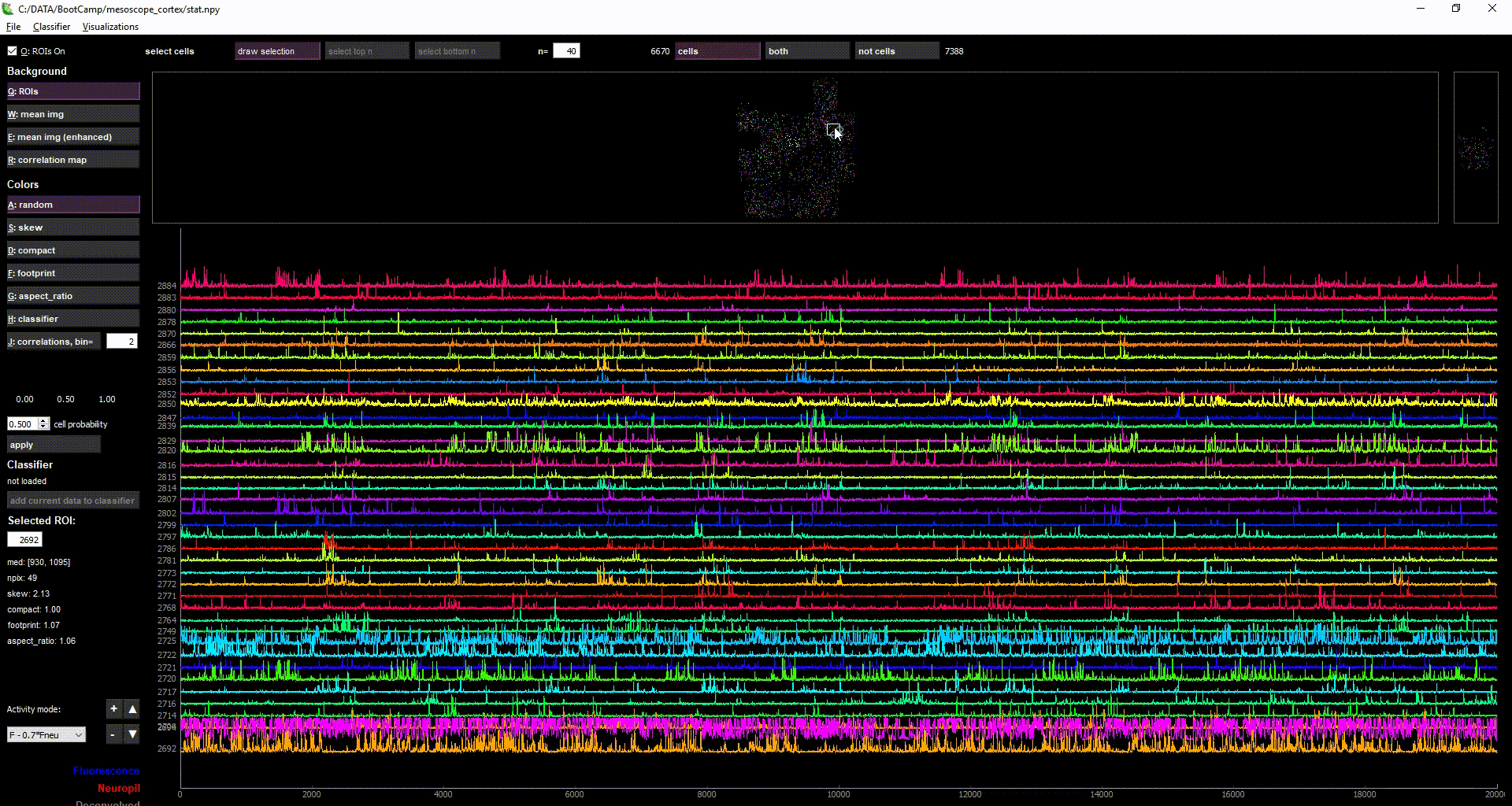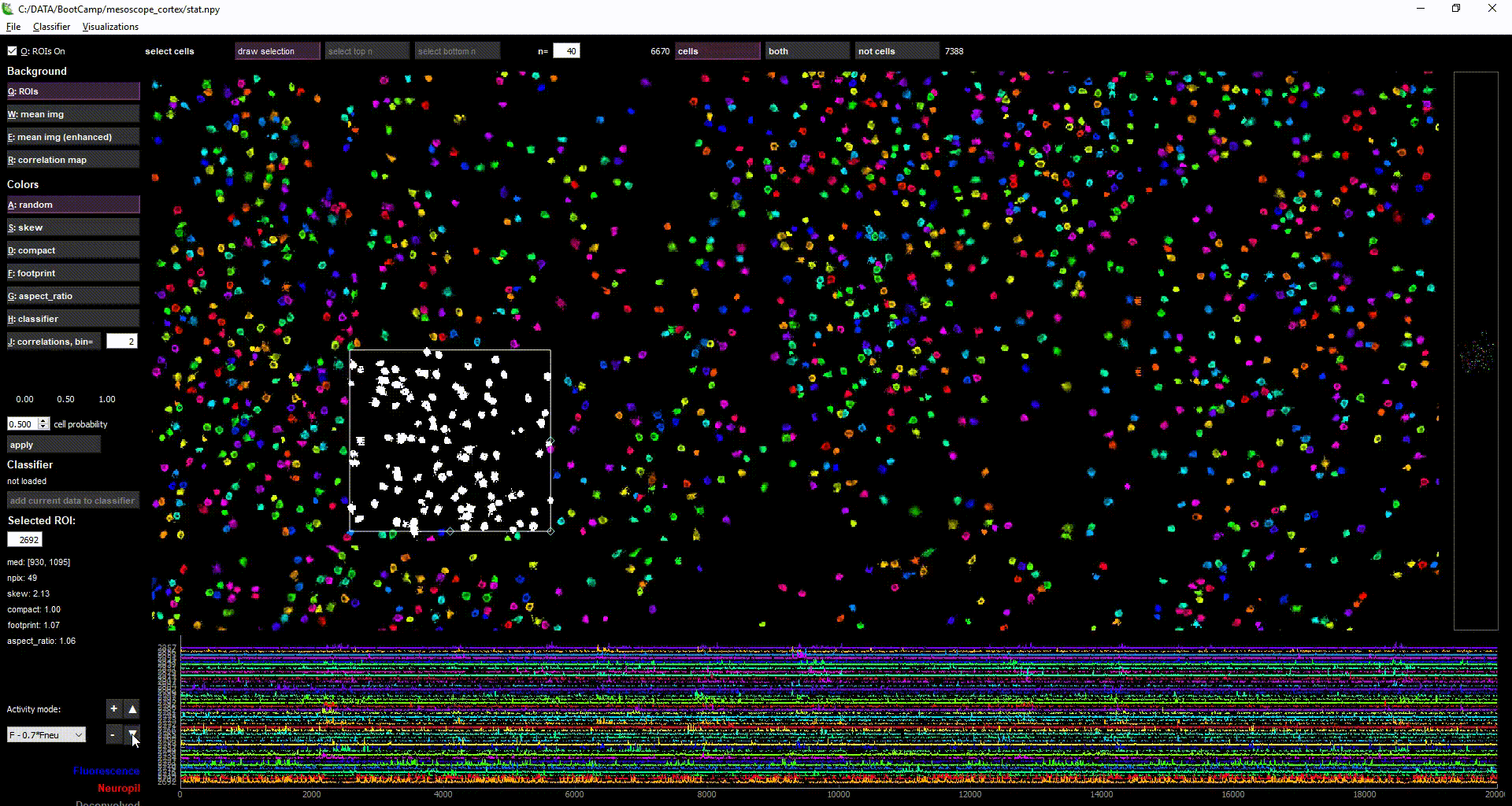 Pachitariu Lab Stringer Lab Python GPL-3.0 Package Calcium imaging Neural recording Google Colab Local installation Napari
Pachitariu Lab Stringer Lab Python GPL-3.0 Package Calcium imaging Neural recording Google Colab Local installation Napari Suite2p
Pipeline for processing two-photon calcium imaging data.
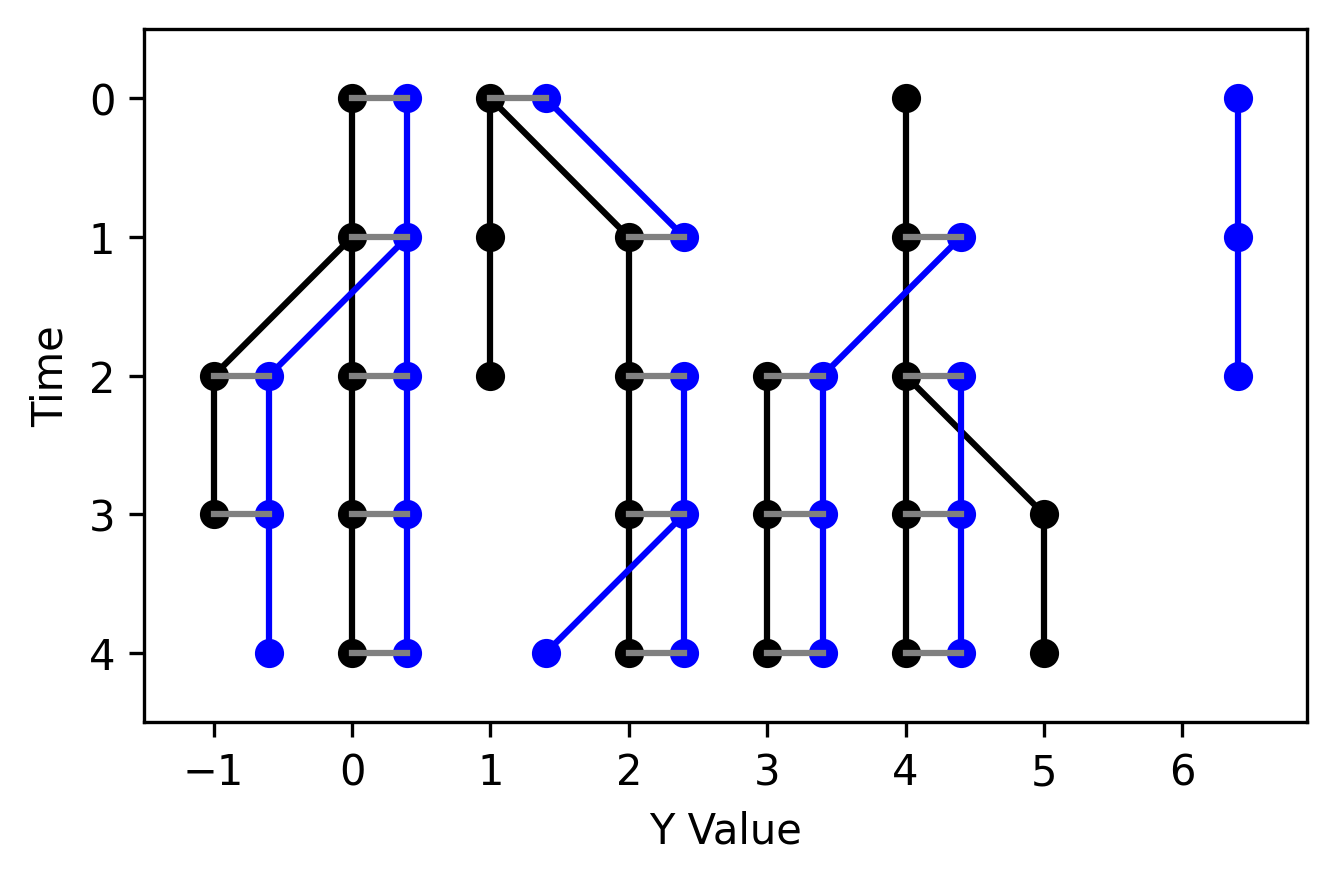 Funke Lab Live Image Tracking Tools Python BSD-3 Clause Package Tracking Local installation
Funke Lab Live Image Tracking Tools Python BSD-3 Clause Package Tracking Local installation Traccuracy
A library for evaluating cell tracking solutions against ground truth annotations.
 Scientific Computing Software Julia BSD-3 Clause Command line application Calcium imaging Electrophysiology Modelling Neural recording HPC cluster Local installation CUDA
Scientific Computing Software Julia BSD-3 Clause Command line application Calcium imaging Electrophysiology Modelling Neural recording HPC cluster Local installation CUDA TrainSpikingNet
train a spiking recurrent neural network
 Branson Lab Scientific Computing Software Javascript Python BSD-3 Clause Framework Package Web application Annotation Cloud Local installation Web browser avi mpeg
Branson Lab Scientific Computing Software Javascript Python BSD-3 Clause Framework Package Web application Annotation Cloud Local installation Web browser avi mpeg Video Annotation Library
A usable and extensible video annotation library for machine learning
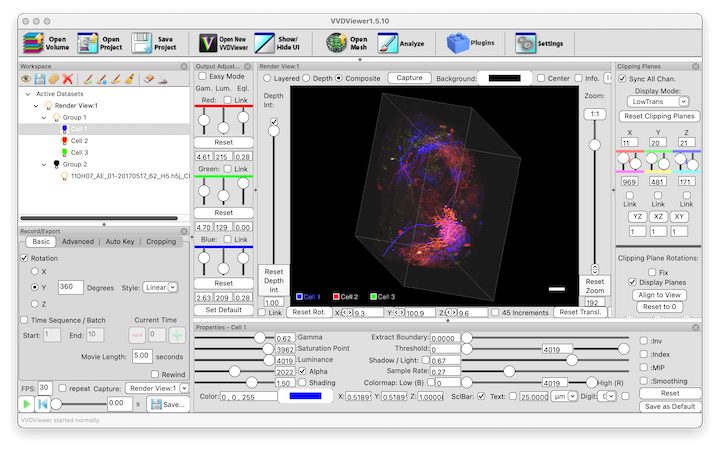 Scientific Computing Software C++ BSD-3 Clause Native application Annotation Confocal light microscopy (LM) Electron microscopy (EM) Expansion microscopy (ExM) Local installation N5 OME-Zarr SWC TIFF ZARR Zeiss CZI
Scientific Computing Software C++ BSD-3 Clause Native application Annotation Confocal light microscopy (LM) Electron microscopy (EM) Expansion microscopy (ExM) Local installation N5 OME-Zarr SWC TIFF ZARR Zeiss CZI VVDViewer
Interactive 3D viewer/renderer for very large image volumes
x2s3
x2s3 is a service which can be used to provide an S3-compatible interface to any storage system. It powers s3.janelia.org.
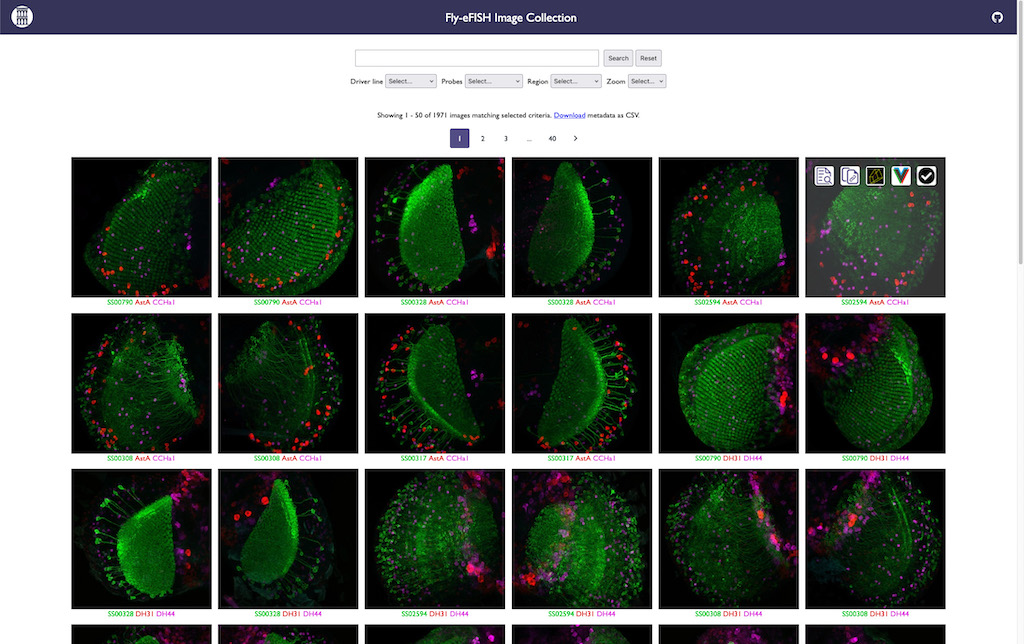 Scientific Computing Software Python BSD-3 Clause Web application Website Confocal light microscopy (LM) Electron microscopy (EM) Infrastructure Web browser
Scientific Computing Software Python BSD-3 Clause Web application Website Confocal light microscopy (LM) Electron microscopy (EM) Infrastructure Web browser Zarrcade
Zarrcade creates searchable web-based galleries of OME-Zarr images.